Many DNA damaging anti-cancer drugs cause replication-associated DNA damage that kill cancer cells. This is an effective way of treating cancer, but the problem is that also normal cells are damaged. Our strategy is to exploit the high level of DNA damage in cancer cells and prevent the repair of these lesions. Using DNA repair inhibitors, we can selectively introduce toxic DNA damage to cancer cells.
The MTH1 Story
High level of DNA damage in cancer – a novel target
It was for a long time well established that cancer cells had a high background level of markers of DNA damage such as γH2AX, which could be related to genetic instability in cancer. In the very same issue of Nature that we demonstrated the BRCA-PARP concept the labs of Jiri Bartek, Thanos Halazonetis and Vassilis Gorgoulis showed that the high level of DNA damage in cancers triggered the p53 tumour barrier, preventing the cancers to grow out. They contacted us and were on track that oncogenes could cause replication stress as a general mechanism to cause DNA damage and senescence in cancer. Natalia Issaeva in the lab helped them validate the replication stress and we published this soon after in Nature.
These data really demonstrated that cancers have a high load of damage at replication forks, similar to that caused by chemotherapy. We were playing with the thought that what if we can stop the repair of these chemotherapy-like DNA damage, then we can find a treatment that can kill the cancer cells and not kill normal cells. This was not a unique idea in our lab and now there was a race to find the way how to selectively kill cancer cells owing to high load of ‘replication stress’. Here, Oscar Fernandez-Capetillo and others did a fantastic job and demonstrated that cancer cells became dependent on the ATR and Chk1 kinases for survival and that they can be targeted by small molecules.
Birth of MTH1 as a target for cancer treatment
The ATR concept was interesting, but both ATR and Chk1 are essential proteins and inhibitors may have off-target effects on normal cells. Ideally, we would want to have a DNA repair protein that is non-essential, and that is required to repair the replication damage specifically occurring in cancer cells. In our lab, Cecilia Lundin assembled an siRNA library consisting of about 200 DNA repair genes that we used for screening. MTH1 was one of the hits coming out of the screens and what really caught our attention was how interesting this enzyme was. It really ticked all boxes for a perfect DNA repair target for anti-cancer treatments and the publications in the literature supported the idea that damage on dNTP pool is very important in cancer. Helge Gad in the lab set out to validate the concept.
At the time our lab was a basic molecular biology lab, but I was really keen to pursue this as a target. MTH1 belonged to the Nudix hydrolase family, and the biochemical assays described used HPLC, which is not suitable for screening compound libraries.
Pyrophosphate is one of the products released from the reaction catalysed by MTH1 and then I came to think about my father’s second cousin, Pål Nyrén, who when bicycling came up with the idea to exploit the release of pyrophosphate as a way of sequencing (this became pyrosequencing). I thought that we could use the same reaction as an effective assay for MTH1 screening, which worked very well.
At this time, we met up with the Chemical Biology team with Thomas Lundbäck and Annika Jenmalm Jensen, who were just in the process of starting up a core facility with equipment donated by Biovitrum AB. They helped with improving the assay to allow screening of larger libraries. To obtain a real MTH1 inhibitor, we would need more chemistry and I hired two fantastic medicinal chemists Martin Scobie and Tobias Koolmeister and placed them at the organic chemistry department at Stockholm University. It is very unusual for a molecular biology lab to hire its own medicinal chemists but turned out fantastic and the two of them really made a huge effort into optimising the hits into potent MTH1 inhibitors.
The open innovation approach
I managed to persuade Camilla Gokturk, who runs a company In vivo Design AB, to start working with us part time to do target validation in vivo and eventually PK/PD work. At this stage, I was persuaded by Annika Jenmalm Jensen that I needed a project leader with Drug discovery expertise and a highly skilled project leader, Ulrika Warpman Berglund came onboard the MTH1 project, which made a huge difference. We had the protein and also a potent inhibitor, so we approached Pål Stenmark, a structural biologist at Stockholm University, who quickly solved the crystal structure of MTH1 with the inhibitors. To optimise the compounds, we needed to know pharmacokinetic properties and started collaborating with Per Artursson at Uppsala University. We now had really promising compounds but needed to know their properties in animals. We also needed to measure the compounds in plasma, and we started a nice collaboration with the analytic chemists at Stockholm University, Ingrid Granelli and Alonja. We continued our open innovation approach and even before publications, many many labs around the globe had the MTH1 inhibitors.
For us, the MTH1 project has demonstrated how powerful interdisciplinary collaboration can be and that an open innovation model is preferred.
During our work we were contacted by Kilian Huber and Giulio Superti-Furga in Vienna. They had identified MTH1 when screening for targets of a compound selectively killing cancer cells. As we already had an MTH1 assay up and running, Ann-Sofie Jemth went to Vienna, in an open innovation approach, to test if their compound was indeed an MTH1 inhibitor. It turned out that indeed it was an inhibitor and then we could use all the resources we had to also progress and validate Kilan’s and Giulio’s discovery.
MTH1 story challenged
After publishing the role of MTH1 and MTH1 inhibitors in Nature (Gad and Koolmeister et al., Nature 2014; Huber et al., Nature 2014), many companies “jumped on” the target and developed MTH1 inhibitors. Soon it was found that the target and pharmacology was not as straight forward as we first anticipated. In 2016, three papers published other MTH1 inhibitors that were potent but not cytotoxic (Kuwamura et al., Sci Rep, 2016; Petrocchi et al., Bioorg Med Chem Lett, 2016; Kettle et al., J Med Chem, 2016). It should however be noted that our previously published data could be reproduced when using our tool compounds and siRNA/shRNA primers.
Today we know that our potent and cytotoxic MTH1 inhibitors act via a dual mechanism; i) causing a mitotic arrest via disturbing tubulin and ii) increasing incorporation of oxidized nucleotides (eg 8-oxodG) into DNA resulting in DNA damage, lagging chromosome, mitotic catastrophy and apoptosis. We are presently further investigating biomarkers, mechanism of action of the MTH1 inhibitors as well as the role of MTH1 in cancer.
Novel MTH1 inhibitor under clinical investigation
At Karolinska Institute, we continued to develop our MTH1 inhibitor and eventually Tobias Koolmeister synthesized our clinical candidate, Karonudib (Karolinska NUDT1 Inhibitor). At this stage of the project, we appointed a clinical director, Teresa Sandvall, to help plan for the clinical development of the compound. Together with Dr. Jeffrey Yachnin, head of the clinical Phase 1 trial, KS, we designed the clinical trial protocol. In the fall of 2016, we sent in application to start Phase 1 trial to regulatory authorities (Medical Product Agency, MPA), and obtained approval in the end of 2016. As an academic group obtaining approval from MPA, fulfilling all requirements, to investigate a first-in-class, novel, MTH1 inhibitor, was a huge achievement!
A clinical Phase 1 trial is presently on-going at Karolinska University Hospital investigating safety and tolerability of the compound in advanced cancer patients with solid malignancies. The sponsor for the study is Helleday Foundation.
The mammalian genome has to be constantly copied (replication), read (transcription) and repaired (DNA repair) which poses a great challenge to eukaryotic cells.
We are interested in deciphering the spatio-temporal coordination of these essential mechanisms using state of the art techniques. Our main interest lies in understanding the interplay of the repair of replication-associated DNA damage, which is induced by many anti-cancer drugs, with on-going transcription.
To this end we have identified various new proteins playing a crucial role at the crossroad of replication, repair and transcription. These new insights will help us to understand the molecular mechanisms of how these cellular processes are regulated and coordinated and might in the end lead to the discovery of new concepts for the treatment of cancer and other diseases.
It’s been known for decades that cancer cells are characterized by switching from predominantly using oxidative phosphorylation for energy to using anaerobic glycolysis for growth. Many are exploiting this switch to tailor novel treatments for cancer and with the modern molecular biology techniques available today we can now selectively inhibit different protein critically involved in cancer growth and potentially less important to normal cells. An example of targets we are working on is MTHFD2, see our pipeline for details.
There are over thirty nucleoside analogs that have been approved as drugs (by FDA and EMA) and several of these are on the WHO list over essential medicines. Most of these drugs are used as antivirals and in the treatment of cancer but they are also used for other indications like immunosuppression and as antiplatelet drugs.
By targeting proteins involved in nucleotide metabolism we can improve the efficacy of current nucleoside treatments. One example of this strategy is the EMA approval in 2016 of the thymidine phosphorylase inhibitor tipiracil to improve trifluridine treatment of metastatic colorectal cancer. For example, we show targeting of SAMHD1 for improved efficacy of AraC treatments (Herold et al., 2017 Nature Medicine). Also, targeting nucleotide metabolism itself can prove highly effective as a monotherapy-based treatment, based on the aberrant metabolism in disease. Read more here (external link).
We are introducing a completely new approach for treatment of inflammatory diseases based on the cross-sectional research areas of DNA repair and inflammation. We have demonstrated proof of principle showing that therapeutic treatment with our tool compound, an OGG1 inhibitor, TH5487 significantly reduces the inflammation in mice with lung inflammation.
We are introducing a completely new approach for treatment of inflammatory diseases based on the cross-sectional research areas of DNA repair and inflammation. We have demonstrated proof of principle showing that therapeutic treatment with our tool compound, an OGG1 inhibitor, TH5487 significantly reduces the inflammation in mice with lung inflammation.
Upon exposure to inflammatory agents, receptor-ligand interactions in cells cause an elevation of reactive oxygen species (ROS) and DNA damage. The pro-inflammatory immune cells that drive auto-immunity and inflammation suffer from high level of oxidative stress and therefore require specific detoxification enzymes for survival and function. One enzyme of specific interest is OGG1, 8-oxo guanine DNA glycosylase 1. Through binding to promoter regions, enriched in oxidized guanines, this enzyme recruits other proteins such as transcription factors, e.g. NF-ƘB to form complexes, promoting transcription of pro-inflammatory genes. Our OGG1 inhibitor TH5487 prevents OGG1 from binding to DNA and thereby dampening the inflammatory responses in animal models.
For more information about the mechanism of action please refer to the original publication.
We are advancing our leading science on the OGG1 pathway to develop novel therapies for severe lung inflammation diseases with large medical need such as ARDS, COPD, severe non allergic asthma, and idiopathic pulmonary fibrosis (IPF).
The Helleday lab is also open for collaborations and partnership to enable development of OGG1 inhibitors as potential anti-inflammatory therapies. Please contact Thomas Helleday directly for further information.
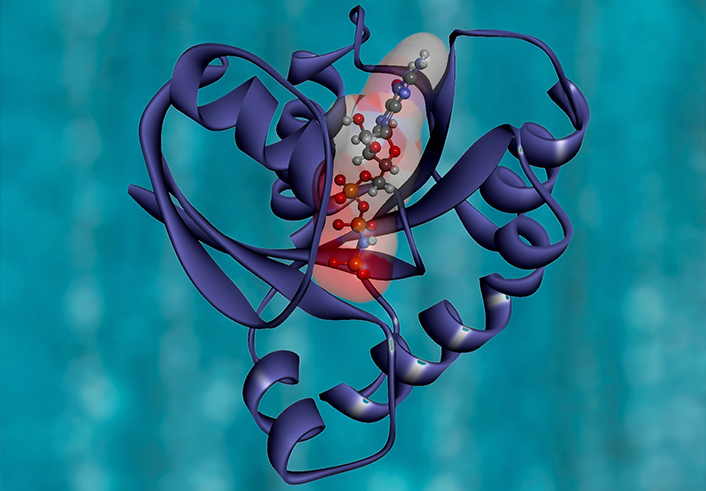
Left: Gallery illustrating the binding mode of 8-oxoguanine modified double stranded DNA to OGG1 (PDB: 1EBM); red to blue color coding of the OGG1 protein indicates a strong positive electrostatic potential of the binding site (blue = positive, red = negative); Later the gallery shows the crystal structure of OGG1 with TH5675, confirming targeting of the positively charged active site; Right: A video generated to demonstrate inhibitor binding and replacement of DNA as a ligand.
Lead generation of OGG1 inhibitors beyond TH5487
Initially, CBK149850 (45% inhibition at 10 µM) was identified as a starting point for lead generation from a high-throughput screen of 17,940 compounds. Interrogation of the structure-activity relationships (SARs) by systematic modification of CBK149850 yielded TH5487 as an initial lead molecule. While substantially more potent (IC50 = 342 nM), obtaining co-crystal structures proved very challenging, due to the limited solubility of both the inhibitors and the protein construct used. In the absence of adequate structural information the SAR of TH5487 was further explored, and collectively over 700 structural analogues were synthesized and tested biochemically. A real breakthrough came when a crystal structure was obtained of mouse OGG1 in complex with TH5675. The mouse homologue of OGG1 has a nearly identical active site but is more soluble due to solubilizing mutations at the protein surface. Furthermore, changing from TH5487 to the less potent but substantially more soluble TH5675 yielded crystals clearly showing bound ligand in the protein active site. To our surprise TH5675 bound with the hydrophobic p-iodophenyl moiety inserted deeply into the 8-oxoguanine pocket, while the benzimidazolone moiety was oriented outward and partially solvent-exposed. With this detailed atomic map at hand, tailored modifications could be made to further improve solubility and metabolic stability while simultaneously improving potency to low nanomolar digits and below. This is illustrated in the figures of the gallery on the left side, where a representative set of OGG1 inhibitors from the TH5487 series are showing progression from TH5487 towards optimal lipophilic ligand efficiency (LLE*) as indicated by the blue region. The second figure further demonstrates the benefits of structure based drug design, as compounds of the TH5487 series synthesized after access to the high resolution crystal structure are highlighted in green and those from before in red.
* for a review see: Leeson PD, Empfield JR (2010) Reducing the Risk of Drug Attrition Associated with Physicochemical Properties. Annu Rev Med Chem 45, 393-407.
This work was funded by the National Institute of Allergic and Infectious Diseases NIAID/AI062885 (I.B.), The Faculty of Medicine at the Norwegian University of Science and Technology and the Central Norway Regional Health Authority (A.S. and H.E.K., project no. 46056921), Svanhild and Arne Must’s Fund for Medical Research (A.S. and H.E.K.), Vinnova (A.C.-K and T.H.), the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 722729 (B.M.F.H. and T.H.), the European Research Council (T.H. TAROX Programme), The Knut and Alice Wallenberg Foundation and the Swedish Foundation for Strategic Research (T.H. and P.S.), Swedish Research Council (T.H. and P.S.), Swedish Cancer Society (T.H. and P.S.), the Swedish Children’s Cancer Foundation (T.H.), the Swedish Pain Relief Foundation (T.H.), and the Torsten and Ragnar Söderberg Foundation (T.H.). This project received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme grant agreement No 957495 DOIFF.
In recent years, the improvement of (bio)chemical reactions has been awarded two noble prizes in chemistry. In both occasions, for enzyme engineering by directed evolution in 2018 and for organocatalysis in 2021, the award acknowledged the tremendous industrial and methodological increase in process efficiency, synthetic possibilities and product yields.
We now have published an initial answer to these challenges in a recent manuscript in the Journal Science.
While these approaches cannot be overrated in their applications, the technologies have not yet reached their full potential in the field of medicine. On the one hand, organocatalysis in living cells would either be protein independent or taking place in the active site of enzymes, with the latter requiring small molecule activation to exist as an orthosteric technology. On the other hand, enzymatic mutations to improve functions within cells would require either short-term translation from for example an RNA-vaccine or permanent and legally challenging CRISPR/Cas methods.
AC50 vs. IC50: TH10785 activates OGG1 function dose-dependently. The assay principle of the OGG1 biochemical assay releases a fluorophore after enzymatic excision of an 8-oxo-purine base. This can be coupled with APE1 for increased turnover or left incubating for slow turnover. Due to OGG1’s rudimentary AP-lyase function the latter version will still lead to full conversion after, just after a longer time frame. These two versions, with and without APE1, are suitable to either identify OGG1 inhibitors (w/ APE1, TH5487) or OGG1 activators (w/o APE1, TH10785). TH10785 has an AC50 of around 700 nM.
Reaction products of mono- and β,δ-bifunctional DNA Glycosylases have distinct repair pathways. Due to its weak AP-lyase function, OGG1 acts as a monofunctional DNA glycosylase in cells. Thus, reaction products are dependent on APE1. Other members of the DNA glycosylase family of enzymes possess β,δ-eliminating properties which require the action of PNKP1 instead. The OGG1 activator TH10785 removes protons from the DNA sugar-backbone, inducing an artificial PNKP1 dependency. At the same time, this reaction is so efficient, that 8-oxo Guanine lesions are neglected and abasic sites are preferred. In cells, the amount of abasic sites dwarfs those of 8-oxo Guanine, allowing TH10785 to be an efficient DNA Glycosylase-AP Lyase switch. In addition, and in contrast to inhibitors where large concentrations are required, low concentrations of activator may be efficient to double or triple the enzymatic function.
We now have published an initial answer to these challenges in a recent manuscript in the Journal Science. We describe the mode of action of the first chemist-synthesized organocatalyst that acts within a proteins active site. The DNA repair enzyme OGG1 or 8-oxo Guanine DNA Glycosylase 1 is boosted with regard to an originally rudimentary function. The increased AP-lyase function reprograms substrate preference from 8-oxo Guanine to abasic sites and further creates a new reaction product. This allows for an increase in oxidative DNA damage repair initiation. At the same, however, this accumulates downstream repair intermediates which are potentially lethal to cells with defective repair systems. When using different classes of the organocatalysts a rational overload or rescue of DNA damage may be achieved. In cells this has shown to work with PNKP1 inhibition. Here, DNA damage directed along the axis 8-oxo Guanine/abasic sites/single strand breaks/double strand breaks sensitizes for defects in DNA damage response. Read our full story here or reach out for more information and collaborate with us. We will continue to build on this discovery and develop workflows for broad development of enzyme targeted organocatalysts – which we have termed Amplizymes.
Situated centrally between all teams, the platform connects the streams of this new technology and establishes the necessary protocols. We are active within the data-driven life science where our platform of choice KNIME enables us to identify future enzyme candidates or target families. Based on a number of descriptors such as enzymatic reactions, substrate scope and pocket size we then rationalize the production of mutants and wild-type enzyme with the biochemistry team. Computational methods are also used to assess the suitability of a compound to perform organocatalysis. Basic determination of pKa, protein-ligand-interactions and functional group tolerance using docking are complemented with quantum-mechanical approaches to identify reaction pathways and perform molecular dynamics calculations. The latter is conducted in collaboration with the Himo lab at Stockholm University. A cornerstone, (Bio)conjugate chemistry uses (glyco)peptides, fluorophores, cleavable linkers and targeting moieties to control delivery, release and potency in cell-compartments which we investigate with the basic science team. Synthesis of Amplizymes expands classical medicinal chemistry, demanding a catalyzing atom center. Here, the necessary method development covers protection group, heterocyclic, photocatalytic and flow chemistry.
Together with biochemistry and in-vitro pharmacology teams, we assess target engagement, potency, substrate scope and kinetic parameter within enzymatic activity assays. Further, to enable substrate and product identification we drive the development of fluorescent, HPLC and gel-based assays for our in-house platforms on DNA glycosylases and the NUDIXes. The in-vivo pharmacology team and UDOPP inform for ADMET properties during the optimization of suitable compounds. We collaborate with the Stenmark lab (Stockholm University) for X-ray structures, the Hertweck lab (Leibniz Institute Jena) for mass spectrometry and the de Vega lab (CBM Severo Ochoa Madrid) for in vitro reconstitution of DNA repair pathways. Translationally we are engaged with the Stolz lab (Goethe University Frankfurt) for autophagy and cellular health screens, the Perona lab for Telomere biology (Alberto Sols, Madrid) and the Research Institutes of Sweden in Södertälje and SINTEF in Trondheim for industrial use of our technology.